Background
Kliegl et al. (2015 ) is a follow-up to Kliegl et al. (2011 ) (see also script kwdyz11.qmd) from an experiment looking at a variety of effects of visual cueing under four different cue-target relations (CTRs). In this experiment two rectangles are displayed (1) in horizontal orientation , (2) in vertical orientation, (3) in left diagonal orientation, or in (4) right diagonal orientation relative to a central fixation point. Subjects react to the onset of a small or a large visual target occurring at one of the four ends of the two rectangles. The target is cued validly on 70% of trials by a brief flash of the corner of the rectangle at which it appears; it is cued invalidly at the three other locations 10% of the trials each. This implies a latent imbalance in design that is not visible in the repeated-measures ANOVA, but we will show its effect in the random-effect structure and conditional modes.
There are a couple of differences between the first and this follow-up experiment, rendering it more a conceptual than a direct replication. First, the original experiment was carried out at Peking University and this follow-up at Potsdam University. Second, diagonal orientations of rectangles and large target sizes were not part of the design of Kliegl et al. (2011 ) .
We specify three contrasts for the four-level factor CTR that are derived from spatial, object-based, and attractor-like features of attention. They map onto sequential differences between appropriately ordered factor levels. Replicating Kliegl et al. (2011 ) , the attraction effect was not significant as a fixed effect, but yielded a highly reliable variance component (VC; i.e., reliable individual differences in positive and negative attraction effects cancel the fixed effect). Moreover, these individual differences in the attraction effect were negatively correlated with those in the spatial effect.
This comparison is of interest because a few years after the publication of Kliegl et al. (2011 ) , the theoretically critical correlation parameter (CP) between the spatial effect and the attraction effect was determined as the source of a non-singular LMM in that paper. The present study served the purpose to estimate this parameter with a larger sample and a wider variety of experimental conditions.
Here we also include two additional experimental manipulations of target size and orientation of cue rectangle. A similar analysis was reported in the parsimonious mixed-model paper (Bates et al., 2015 ) ; it was also used in a paper of GAMMs (Baayen et al., 2017 ) . Data and R scripts of those analyses are also available in R-package RePsychLing .
The analysis is based on log-transformed reaction times lrt, indicated by a boxcox() check of model residuals.
In this vignette we focus on the reduction of model complexity. And we start with a quote:
“Neither the [maximal] nor the [minimal] linear mixed models are appropriate for most repeated measures analysis. Using the [maximal] model is generally wasteful and costly in terms of statiscal power for testing hypotheses. On the other hand, the [minimal] model fails to account for nontrivial correlation among repeated measurements. This results in inflated [T]ype I error rates when non-negligible correlation does in fact exist. We can usually find middle ground, a covariance model that adequately accounts for correlation but is more parsimonious than the [maximal] model. Doing so allows us full control over [T]ype I error rates without needlessly sacrificing power.”
Stroup, W. W. (2012, p. 185). Generalized linear mixed models: Modern concepts, methods and applica?ons. CRC Press, Boca Raton.
Packages
Code
using Arrow using AlgebraOfGraphics using AlgebraOfGraphics : densityusing BoxCox using CairoMakie using CategoricalArrays using Chain using DataFrameMacros using DataFrames using MixedModels using MixedModelsMakie using ProgressMeter using Random using SMLP2023 : datasetusing StatsBase ijulia_behavior (: clear)activate! (; type = "svg" )
Read data, compute and plot means
= DataFrame (dataset (: kkl15))describe (dat)
1
Subj
S001
S147
0
String
2
CTR
dod
val
0
String
3
rt
293.147
150.22
276.594
749.481
0
Float32
4
cardinal
cardinal
diagonal
0
String
5
size
big
small
0
String
= combine (groupby (dat, [: Subj, : CTR]),=> : n,: rt => mean => : rt_m,: rt => (c -> mean (log, c)) => : lrt_m,= categorical (dat_subj.CTR, levels= levels (dat.CTR))describe (dat_subj)
1
Subj
S001
S147
0
String
2
CTR
val
dod
0
CategoricalValue{String, UInt32}
3
n
156.294
49
64.0
448
0
Int64
4
rt_m
308.223
208.194
304.862
584.71
0
Float32
5
lrt_m
5.6908
5.33226
5.69848
6.36141
0
Float32
Code
boxplot (=: horizontal,= true ,= (;= (1 : 4 ,"valid cue" ,"same obj/diff pos" ,"diff obj/same pos" ,"diff obj/diff pos" ,= (; resolution= (800 , 300 )),
Mean of log reaction times for four cue-target relations. Targets appeared at (a) the cued position (valid) in a rectangle, (b) in the same rectangle cue, but at its other end, (c) on the second rectangle, but at a corresponding horizontal/vertical physical distance, or (d) at the other end of the second rectangle, that is \(\sqrt{2}\) of horizontal/vertical distance diagonally across from the cue, that is also at larger physical distance compared to (c).
We remove the outlier subject and replot, but we model the data points in dat and check whether this subject appears as an outlier in the caterpillar plot of conditional modes.
Code
let dat_subj = filter (r -> r.rt_m < 510 , dat_subj)boxplot (=: horizontal,= true ,= (;= (1 : 4 ,"valid cue" ,"same obj/diff pos" ,"diff obj/same pos" ,"diff obj/diff pos" ,= (; resolution= (800 , 300 )),end
Setup of linear mixed model
Contrasts
= Dict (: Subj => Grouping (),: CTR => SeqDiffCoding (; levels= ["val" , "sod" , "dos" , "dod" ]),: cardinal => EffectsCoding (; levels= ["cardinal" , "diagonal" ]),: size => EffectsCoding (; levels= ["big" , "small" ])
Dict{Symbol, StatsModels.AbstractContrasts} with 4 entries:
:CTR => SeqDiffCoding(["val", "sod", "dos", "dod"])
:size => EffectsCoding(nothing, ["big", "small"])
:Subj => Grouping()
:cardinal => EffectsCoding(nothing, ["cardinal", "diagonal"])
= let = @formula rt ~ 1 + CTR * size * cardinal + 1 + CTR * size * cardinal | Subj)fit (MixedModel, form, dat; contrasts)end
Minimizing 3872 Time: 0:00:06 ( 1.59 ms/it)
objective: 599297.6610000695
(Intercept)
308.4078
5.8365
52.84
<1e-99
0.0000
CTR: sod
23.2685
2.5676
9.06
<1e-18
9.9631
CTR: dos
13.0802
1.5433
8.48
<1e-16
5.0118
CTR: dod
2.7724
2.0921
1.33
0.1851
13.1147
size: small
26.4130
5.8365
4.53
<1e-05
54.0095
cardinal: diagonal
6.6749
1.7890
3.73
0.0002
9.7692
CTR: sod & size: small
8.7924
2.5676
3.42
0.0006
19.8731
CTR: dos & size: small
-0.7061
1.5433
-0.46
0.6473
7.2502
CTR: dod & size: small
7.4980
2.0921
3.58
0.0003
8.7387
CTR: sod & cardinal: diagonal
3.6315
1.1022
3.29
0.0010
4.2292
CTR: dos & cardinal: diagonal
1.3344
1.2450
1.07
0.2838
1.7717
CTR: dod & cardinal: diagonal
-0.2247
1.3224
-0.17
0.8651
4.0782
size: small & cardinal: diagonal
2.0502
1.7890
1.15
0.2518
12.9327
CTR: sod & size: small & cardinal: diagonal
-0.7638
1.1022
-0.69
0.4883
3.7198
CTR: dos & size: small & cardinal: diagonal
-0.1556
1.2450
-0.12
0.9005
1.7390
CTR: dod & size: small & cardinal: diagonal
4.1768
1.3224
3.16
0.0016
2.3972
Residual
63.0091
= let = @formula rt ~ 1 + CTR * size * cardinal + 1 + CTR + size + cardinal | Subj)fit (MixedModel, form, dat; contrasts)end
Minimizing 770 Time: 0:00:00 ( 0.25 ms/it)
(Intercept)
308.4074
6.0527
50.95
<1e-99
40.4433
CTR: sod
23.2723
2.4739
9.41
<1e-20
21.2925
CTR: dos
13.1099
1.4621
8.97
<1e-18
7.5060
CTR: dod
2.7140
1.9182
1.41
0.1571
13.7125
size: small
26.3865
6.0527
4.36
<1e-04
38.7600
cardinal: diagonal
6.6506
1.7311
3.84
0.0001
15.6567
CTR: sod & size: small
8.7956
2.4739
3.56
0.0004
CTR: dos & size: small
-0.7230
1.4621
-0.49
0.6209
CTR: dod & size: small
7.4191
1.9182
3.87
0.0001
CTR: sod & cardinal: diagonal
3.6414
0.9210
3.95
<1e-04
CTR: dos & cardinal: diagonal
1.3276
1.2176
1.09
0.2755
CTR: dod & cardinal: diagonal
-0.3142
1.2217
-0.26
0.7970
size: small & cardinal: diagonal
2.0470
1.7311
1.18
0.2370
CTR: sod & size: small & cardinal: diagonal
-0.7611
0.9210
-0.83
0.4086
CTR: dos & size: small & cardinal: diagonal
-0.1334
1.2176
-0.11
0.9128
CTR: dod & size: small & cardinal: diagonal
4.1115
1.2217
3.37
0.0008
Residual
63.1011
Box-Cox
= fit (BoxCoxTransformation, m_max_rt)
= fit (BoxCoxTransformation, m_cpx_rt)
Minimizing 1191 Time: 0:00:00 ( 0.14 ms/it)
Box-Cox transformation
estimated λ: -0.7062
resultant transformation:
y^-0.7 - 1
------------
-0.7
boxcoxplot (bc2; conf_level= 0.95 )
Clear evidence for skew. Traditionally, we used log transforms for reaction times. even stronger than log. We stay with log for now. Could try 1/sqrt(rt).
Maximum LMM
This is the maximum LMM for the design.
= let = @formula log (rt) ~ 1 + CTR * size * cardinal + 1 + CTR * size * cardinal | Subj)fit (MixedModel, form, dat; contrasts)end
Minimizing 8282 Time: 0:00:13 ( 1.63 ms/it)
objective: -24691.24618930951
(Intercept)
5.6911
0.0174
327.65
<1e-99
0.0000
CTR: sod
0.0744
0.0078
9.52
<1e-20
0.0096
CTR: dos
0.0408
0.0047
8.75
<1e-17
0.0135
CTR: dod
0.0018
0.0056
0.31
0.7553
0.0233
size: small
0.0921
0.0174
5.30
<1e-06
0.1607
cardinal: diagonal
0.0205
0.0052
3.91
<1e-04
0.0261
CTR: sod & size: small
0.0244
0.0078
3.12
0.0018
0.0671
CTR: dos & size: small
-0.0054
0.0047
-1.15
0.2492
0.0231
CTR: dod & size: small
0.0183
0.0056
3.25
0.0011
0.0320
CTR: sod & cardinal: diagonal
0.0101
0.0032
3.12
0.0018
0.0074
CTR: dos & cardinal: diagonal
0.0046
0.0037
1.24
0.2149
0.0039
CTR: dod & cardinal: diagonal
-0.0054
0.0039
-1.39
0.1648
0.0082
size: small & cardinal: diagonal
0.0042
0.0052
0.80
0.4264
0.0395
CTR: sod & size: small & cardinal: diagonal
-0.0032
0.0032
-0.98
0.3287
0.0135
CTR: dos & size: small & cardinal: diagonal
-0.0005
0.0037
-0.14
0.8925
0.0033
CTR: dod & size: small & cardinal: diagonal
0.0111
0.0039
2.85
0.0044
0.0082
Residual
0.1902
only (MixedModels.PCA (m_max))
Principal components based on correlation matrix
(Intercept) … . . . . .
CTR: sod . . . . .
CTR: dos . . . . .
CTR: dod . . . . .
size: small . . . . .
CTR: sod & size: small … . . . . .
CTR: dos & size: small . . . . .
CTR: dod & size: small . . . . .
cardinal: diagonal . . . . .
CTR: sod & cardinal: diagonal . . . . .
CTR: dos & cardinal: diagonal … . . . . .
CTR: dod & cardinal: diagonal 1.0 . . . .
size: small & cardinal: diagonal 0.03 1.0 . . .
CTR: sod & size: small & cardinal: diagonal 0.44 -0.02 1.0 . .
CTR: dos & size: small & cardinal: diagonal 0.75 0.36 0.1 1.0 .
CTR: dod & size: small & cardinal: diagonal … 0.28 -0.05 0.67 -0.13 1.0
Normalized cumulative variances:
[0.3, 0.577, 0.7187, 0.8235, 0.9131, 0.9482, 0.9806, 0.9983, 0.9997, 0.9999, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0]
Component loadings
… PC13 PC14 PC15 PC16
(Intercept) -0.0 -0.0 0.0 -1.0
CTR: sod -0.03 0.07 -0.02 0.0
CTR: dos 0.41 0.2 0.12 -0.0
CTR: dod -0.02 -0.25 0.29 0.0
size: small … 0.06 -0.26 -0.47 -0.0
CTR: sod & size: small 0.15 0.49 -0.11 -0.0
CTR: dos & size: small 0.41 0.36 0.24 -0.0
CTR: dod & size: small -0.16 -0.35 0.47 0.0
cardinal: diagonal -0.3 0.16 0.1 0.0
CTR: sod & cardinal: diagonal … 0.11 -0.06 0.04 -0.0
CTR: dos & cardinal: diagonal -0.04 -0.33 -0.17 0.0
CTR: dod & cardinal: diagonal -0.32 0.11 -0.05 0.0
size: small & cardinal: diagonal -0.37 0.26 0.11 -0.0
CTR: sod & size: small & cardinal: diagonal 0.21 -0.14 0.16 -0.0
CTR: dos & size: small & cardinal: diagonal … 0.35 -0.2 -0.4 0.0
CTR: dod & size: small & cardinal: diagonal -0.32 0.22 -0.39 0.0
Subj
(Intercept)
0.00000000
0.00000000
CTR: sod
0.00009280
0.00963318
+NaN
CTR: dos
0.00018297
0.01352646
+NaN
-0.01
CTR: dod
0.00054215
0.02328402
+NaN
+0.51
-0.35
size: small
0.02583099
0.16072022
+NaN
+0.90
+0.19
+0.67
CTR: sod & size: small
0.00449651
0.06705605
+NaN
+0.10
+0.32
+0.55
+0.50
CTR: dos & size: small
0.00053188
0.02306260
+NaN
+0.26
-0.74
+0.66
+0.19
-0.09
CTR: dod & size: small
0.00102251
0.03197673
+NaN
+0.40
+0.45
+0.49
+0.68
+0.70
+0.15
cardinal: diagonal
0.00068321
0.02613836
+NaN
+0.14
-0.55
+0.06
-0.07
-0.23
+0.14
-0.49
CTR: sod & cardinal: diagonal
0.00005543
0.00744533
+NaN
+0.25
-0.30
-0.25
-0.01
-0.43
+0.05
-0.32
+0.25
CTR: dos & cardinal: diagonal
0.00001502
0.00387533
+NaN
-0.79
+0.30
-0.18
-0.48
+0.40
-0.31
+0.02
-0.34
-0.42
CTR: dod & cardinal: diagonal
0.00006782
0.00823543
+NaN
+0.35
-0.55
+0.34
+0.14
-0.36
+0.83
+0.17
+0.02
+0.24
-0.59
size: small & cardinal: diagonal
0.00156110
0.03951070
+NaN
-0.02
+0.06
+0.09
+0.07
-0.06
+0.24
+0.09
-0.49
+0.10
+0.29
+0.03
CTR: sod & size: small & cardinal: diagonal
0.00018092
0.01345048
+NaN
+0.28
-0.08
+0.15
+0.26
+0.04
+0.28
+0.35
-0.21
-0.13
-0.33
+0.44
-0.02
CTR: dos & size: small & cardinal: diagonal
0.00001090
0.00330194
+NaN
+0.09
-0.75
+0.38
-0.06
-0.36
+0.91
-0.10
+0.26
+0.20
-0.23
+0.75
+0.36
+0.10
CTR: dod & size: small & cardinal: diagonal
0.00006649
0.00815442
+NaN
+0.23
+0.34
+0.37
+0.47
+0.55
+0.17
+0.87
-0.56
-0.43
-0.01
+0.28
-0.05
+0.67
-0.13
Residual
0.03617587
0.19019955
Reduction strategy 1
Zero-correlation parameter LMM (1)
Force CPs to zero.
= let = @formula log (rt) ~ 1 + CTR * size * cardinal + zerocorr (1 + CTR * size * cardinal | Subj)fit (MixedModel, form, dat; contrasts)end
Minimizing 637 Time: 0:00:00 ( 0.32 ms/it)
objective: -24588.607469898558
(Intercept)
5.6910
0.0173
329.51
<1e-99
0.0151
CTR: sod
0.0744
0.0077
9.66
<1e-21
0.0109
CTR: dos
0.0409
0.0045
9.09
<1e-19
0.0240
CTR: dod
0.0015
0.0053
0.29
0.7749
0.0299
size: small
0.0921
0.0173
5.33
<1e-07
0.1591
cardinal: diagonal
0.0204
0.0052
3.92
<1e-04
0.0312
CTR: sod & size: small
0.0244
0.0077
3.17
0.0015
0.0657
CTR: dos & size: small
-0.0053
0.0045
-1.19
0.2344
0.0010
CTR: dod & size: small
0.0181
0.0053
3.39
0.0007
0.0196
CTR: sod & cardinal: diagonal
0.0101
0.0033
3.03
0.0025
0.0172
CTR: dos & cardinal: diagonal
0.0046
0.0037
1.26
0.2092
0.0000
CTR: dod & cardinal: diagonal
-0.0055
0.0037
-1.49
0.1358
0.0033
size: small & cardinal: diagonal
0.0041
0.0052
0.79
0.4291
0.0354
CTR: sod & size: small & cardinal: diagonal
-0.0032
0.0033
-0.95
0.3427
0.0008
CTR: dos & size: small & cardinal: diagonal
-0.0005
0.0037
-0.13
0.8945
0.0000
CTR: dod & size: small & cardinal: diagonal
0.0109
0.0037
2.94
0.0033
0.0010
Residual
0.1903
only (MixedModels.PCA (m_zcp1))
Principal components based on correlation matrix
(Intercept) … . . . . . . .
CTR: sod . . . . . . .
CTR: dos . . . . . . .
CTR: dod . . . . . . .
size: small . . . . . . .
CTR: sod & size: small … . . . . . . .
CTR: dos & size: small . . . . . . .
CTR: dod & size: small . . . . . . .
cardinal: diagonal . . . . . . .
CTR: sod & cardinal: diagonal 1.0 . . . . . .
CTR: dos & cardinal: diagonal … 0.0 0.0 . . . . .
CTR: dod & cardinal: diagonal 0.0 0.0 1.0 . . . .
size: small & cardinal: diagonal 0.0 0.0 0.0 1.0 . . .
CTR: sod & size: small & cardinal: diagonal 0.0 0.0 0.0 0.0 1.0 . .
CTR: dos & size: small & cardinal: diagonal 0.0 0.0 0.0 0.0 0.0 0.0 .
CTR: dod & size: small & cardinal: diagonal … 0.0 0.0 0.0 0.0 0.0 0.0 1.0
Normalized cumulative variances:
[0.0714, 0.1429, 0.2143, 0.2857, 0.3571, 0.4286, 0.5, 0.5714, 0.6429, 0.7143, 0.7857, 0.8571, 0.9286, 1.0, 1.0, 1.0]
Component loadings
… PC13 PC14 PC15 PC16
(Intercept) 0.0 0.0 0.0 0.0
CTR: sod 0.0 0.0 0.0 0.0
CTR: dos 0.0 0.0 0.0 0.0
CTR: dod 0.0 0.0 0.0 0.0
size: small … 0.0 0.0 0.0 0.0
CTR: sod & size: small 0.0 0.0 0.0 0.0
CTR: dos & size: small 0.0 0.0 0.0 0.0
CTR: dod & size: small 1.0 0.0 0.0 0.0
cardinal: diagonal 0.0 1.0 0.0 0.0
CTR: sod & cardinal: diagonal … 0.0 0.0 0.0 0.0
CTR: dos & cardinal: diagonal 0.0 0.0 NaN 0.0
CTR: dod & cardinal: diagonal 0.0 0.0 0.0 0.0
size: small & cardinal: diagonal 0.0 0.0 0.0 0.0
CTR: sod & size: small & cardinal: diagonal 0.0 0.0 0.0 0.0
CTR: dos & size: small & cardinal: diagonal … 0.0 0.0 0.0 NaN
CTR: dod & size: small & cardinal: diagonal 0.0 0.0 0.0 0.0
Subj
(Intercept)
0.000228119
0.015103595
CTR: sod
0.000118184
0.010871228
.
CTR: dos
0.000577145
0.024023842
.
.
CTR: dod
0.000894364
0.029905924
.
.
.
size: small
0.025310609
0.159093081
.
.
.
.
CTR: sod & size: small
0.004316747
0.065701959
.
.
.
.
.
CTR: dos & size: small
0.000000926
0.000962365
.
.
.
.
.
.
CTR: dod & size: small
0.000383055
0.019571794
.
.
.
.
.
.
.
cardinal: diagonal
0.000972318
0.031182012
.
.
.
.
.
.
.
.
CTR: sod & cardinal: diagonal
0.000294448
0.017159499
.
.
.
.
.
.
.
.
.
CTR: dos & cardinal: diagonal
0.000000000
0.000000000
.
.
.
.
.
.
.
.
.
.
CTR: dod & cardinal: diagonal
0.000010975
0.003312812
.
.
.
.
.
.
.
.
.
.
.
size: small & cardinal: diagonal
0.001254094
0.035413184
.
.
.
.
.
.
.
.
.
.
.
.
CTR: sod & size: small & cardinal: diagonal
0.000000719
0.000847671
.
.
.
.
.
.
.
.
.
.
.
.
.
CTR: dos & size: small & cardinal: diagonal
0.000000000
0.000000000
.
.
.
.
.
.
.
.
.
.
.
.
.
.
CTR: dod & size: small & cardinal: diagonal
0.000001038
0.001019064
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
Residual
0.036196631
0.190254123
Reduced zcp LMM
Take out VC for interactions.
= let = @formula log (rt) ~ 1 + CTR * size * cardinal + zerocorr (1 + CTR + size + cardinal | Subj)fit (MixedModel, form, dat; contrasts)end
(Intercept)
5.6911
0.0173
329.28
<1e-99
0.1569
CTR: sod
0.0745
0.0077
9.66
<1e-21
0.0666
CTR: dos
0.0409
0.0045
9.09
<1e-19
0.0240
CTR: dod
0.0015
0.0053
0.29
0.7751
0.0357
size: small
0.0920
0.0173
5.33
<1e-06
0.0312
cardinal: diagonal
0.0204
0.0053
3.88
0.0001
0.0476
CTR: sod & size: small
0.0244
0.0077
3.17
0.0015
CTR: dos & size: small
-0.0054
0.0045
-1.20
0.2317
CTR: dod & size: small
0.0181
0.0053
3.39
0.0007
CTR: sod & cardinal: diagonal
0.0101
0.0028
3.64
0.0003
CTR: dos & cardinal: diagonal
0.0046
0.0037
1.25
0.2125
CTR: dod & cardinal: diagonal
-0.0056
0.0037
-1.51
0.1317
size: small & cardinal: diagonal
0.0042
0.0053
0.79
0.4286
CTR: sod & size: small & cardinal: diagonal
-0.0031
0.0028
-1.12
0.2610
CTR: dos & size: small & cardinal: diagonal
-0.0005
0.0037
-0.12
0.9023
CTR: dod & size: small & cardinal: diagonal
0.0109
0.0037
2.95
0.0032
Residual
0.1904
only (MixedModels.PCA (m_zcp1_rdc))
Principal components based on correlation matrix
(Intercept) 1.0 . . . . .
CTR: sod 0.0 1.0 . . . .
CTR: dos 0.0 0.0 1.0 . . .
CTR: dod 0.0 0.0 0.0 1.0 . .
size: small 0.0 0.0 0.0 0.0 1.0 .
cardinal: diagonal 0.0 0.0 0.0 0.0 0.0 1.0
Normalized cumulative variances:
[0.1667, 0.3333, 0.5, 0.6667, 0.8333, 1.0]
Component loadings
PC1 PC2 PC3 PC4 PC5 PC6
(Intercept) 1.0 0.0 0.0 0.0 0.0 0.0
CTR: sod 0.0 1.0 0.0 0.0 0.0 0.0
CTR: dos 0.0 0.0 1.0 0.0 0.0 0.0
CTR: dod 0.0 0.0 0.0 1.0 0.0 0.0
size: small 0.0 0.0 0.0 0.0 1.0 0.0
cardinal: diagonal 0.0 0.0 0.0 0.0 0.0 1.0
Subj
(Intercept)
0.0246022
0.1568508
CTR: sod
0.0044408
0.0666395
.
CTR: dos
0.0005763
0.0240065
.
.
CTR: dod
0.0012752
0.0357098
.
.
.
size: small
0.0009731
0.0311946
.
.
.
.
cardinal: diagonal
0.0022679
0.0476222
.
.
.
.
.
Residual
0.0362601
0.1904207
Model comparison 1
Let’s compare the three models.
= let = [: m_zcp1_rdc, : m_zcp1, : m_max]= eval .(nms)= MixedModels.likelihoodratiotest (m_zcp1_rdc, m_zcp1, m_max)DataFrame (;= nms, = dof .(mods),= round .(deviance .(mods), digits= 0 ),= round .(aic .(mods),digits= 0 ),= round .(aicc .(mods),digits= 0 ),= round .(bic .(mods),digits= 0 ),= vcat (: ., round .(lrt.tests.deviancediff, digits= 0 )),= vcat (: ., round .(lrt.tests.dofdiff, digits= 0 )),= vcat (: ., round .(lrt.tests.pvalues, digits= 3 ))end
1
m_zcp1_rdc
23
-24558.0
-24512.0
-24512.0
-24308.0
.
.
.
2
m_zcp1
33
-24589.0
-24523.0
-24523.0
-24229.0
30.0
10.0
0.001
3
m_max
153
-24691.0
-24385.0
-24384.0
-23025.0
103.0
120.0
0.872
Parsimonious LMM (1)
Extend zcp-reduced LMM with CPs
= let = @formula log (rt) ~ 1 + CTR * size * cardinal + 1 + CTR + size + cardinal | Subj)fit (MixedModel, form, dat; contrasts)end
Minimizing 801 Time: 0:00:00 ( 0.18 ms/it)
(Intercept)
5.6911
0.0176
323.61
<1e-99
0.1181
CTR: sod
0.0745
0.0076
9.75
<1e-21
0.0660
CTR: dos
0.0408
0.0043
9.42
<1e-20
0.0213
CTR: dod
0.0017
0.0051
0.34
0.7374
0.0326
size: small
0.0921
0.0176
5.23
<1e-06
0.1119
cardinal: diagonal
0.0204
0.0053
3.88
0.0001
0.0476
CTR: sod & size: small
0.0244
0.0076
3.19
0.0014
CTR: dos & size: small
-0.0054
0.0043
-1.24
0.2156
CTR: dod & size: small
0.0181
0.0051
3.55
0.0004
CTR: sod & cardinal: diagonal
0.0101
0.0028
3.64
0.0003
CTR: dos & cardinal: diagonal
0.0046
0.0037
1.25
0.2118
CTR: dod & cardinal: diagonal
-0.0056
0.0037
-1.51
0.1301
size: small & cardinal: diagonal
0.0042
0.0053
0.79
0.4272
CTR: sod & size: small & cardinal: diagonal
-0.0031
0.0028
-1.13
0.2597
CTR: dos & size: small & cardinal: diagonal
-0.0005
0.0037
-0.13
0.8997
CTR: dod & size: small & cardinal: diagonal
0.0110
0.0037
2.98
0.0029
Residual
0.1904
only (MixedModels.PCA (m_prm1))
Principal components based on correlation matrix
(Intercept) 1.0 . . . . .
CTR: sod 0.64 1.0 . . . .
CTR: dos 0.16 -0.08 1.0 . . .
CTR: dod 0.79 0.6 0.28 1.0 . .
size: small -0.42 -0.16 -0.12 -0.03 1.0 .
cardinal: diagonal 0.05 -0.05 -0.02 0.05 0.08 1.0
Normalized cumulative variances:
[0.414, 0.5967, 0.772, 0.926, 0.9822, 1.0]
Component loadings
PC1 PC2 PC3 PC4 PC5 PC6
(Intercept) -0.6 0.0 0.03 0.12 -0.37 -0.7
CTR: sod -0.5 -0.26 0.31 -0.11 0.76 0.02
CTR: dos -0.16 0.52 -0.72 -0.23 0.34 -0.11
CTR: dod -0.55 -0.18 -0.2 -0.27 -0.4 0.63
size: small 0.25 -0.59 -0.23 -0.66 -0.03 -0.32
cardinal: diagonal -0.0 -0.53 -0.54 0.64 0.12 0.03
Subj
(Intercept)
0.0139568
0.1181388
CTR: sod
0.0043556
0.0659971
+0.64
CTR: dos
0.0004536
0.0212981
+0.16
-0.08
CTR: dod
0.0010640
0.0326186
+0.79
+0.60
+0.28
size: small
0.0125269
0.1119239
-0.42
-0.16
-0.12
-0.03
cardinal: diagonal
0.0022690
0.0476336
+0.05
-0.05
-0.02
+0.05
+0.08
Residual
0.0362661
0.1904365
We note that the critical correlation parameter between spatial (sod) and attraction (dod) is now estimated at .60 – not that close to the 1.0 boundary that caused singularity in Kliegl et al. (2011 ) .
Model comparison 2
= let = [: m_zcp1_rdc, : m_prm1, : m_max]= eval .(nms)= MixedModels.likelihoodratiotest (m_prm1, m_zcp1, m_max)DataFrame (;= nms, = dof .(mods),= round .(deviance .(mods), digits= 0 ),= round .(aic .(mods),digits= 0 ),= round .(aicc .(mods),digits= 0 ),= round .(bic .(mods),digits= 0 ),= vcat (: ., round .(lrt.tests.deviancediff, digits= 0 )),= vcat (: ., round .(lrt.tests.dofdiff, digits= 0 )),= vcat (: ., round .(lrt.tests.pvalues, digits= 3 ))end
1
m_zcp1_rdc
23
-24558.0
-24512.0
-24512.0
-24308.0
.
.
.
2
m_prm1
38
-24615.0
-24539.0
-24539.0
-24201.0
27.0
5.0
0.0
3
m_max
153
-24691.0
-24385.0
-24384.0
-23025.0
76.0
115.0
0.998
Reduction strategy 2
Complex LMM
Take out interaction VCs.
= let = @formula log (rt) ~ 1 + CTR * size * cardinal + 1 + CTR + size + cardinal | Subj)fit (MixedModel, form, dat; contrasts)end
Minimizing 801 Time: 0:00:00 ( 0.19 ms/it)
(Intercept)
5.6911
0.0176
323.61
<1e-99
0.1181
CTR: sod
0.0745
0.0076
9.75
<1e-21
0.0660
CTR: dos
0.0408
0.0043
9.42
<1e-20
0.0213
CTR: dod
0.0017
0.0051
0.34
0.7374
0.0326
size: small
0.0921
0.0176
5.23
<1e-06
0.1119
cardinal: diagonal
0.0204
0.0053
3.88
0.0001
0.0476
CTR: sod & size: small
0.0244
0.0076
3.19
0.0014
CTR: dos & size: small
-0.0054
0.0043
-1.24
0.2156
CTR: dod & size: small
0.0181
0.0051
3.55
0.0004
CTR: sod & cardinal: diagonal
0.0101
0.0028
3.64
0.0003
CTR: dos & cardinal: diagonal
0.0046
0.0037
1.25
0.2118
CTR: dod & cardinal: diagonal
-0.0056
0.0037
-1.51
0.1301
size: small & cardinal: diagonal
0.0042
0.0053
0.79
0.4272
CTR: sod & size: small & cardinal: diagonal
-0.0031
0.0028
-1.13
0.2597
CTR: dos & size: small & cardinal: diagonal
-0.0005
0.0037
-0.13
0.8997
CTR: dod & size: small & cardinal: diagonal
0.0110
0.0037
2.98
0.0029
Residual
0.1904
Zero-correlation parameter LMM (2)
Take out interaction VCs.
= let = @formula log (rt) ~ 1 + CTR * size * cardinal + zerocorr (1 + CTR + size + cardinal | Subj)fit (MixedModel, form, dat; contrasts)end
(Intercept)
5.6911
0.0173
329.28
<1e-99
0.1569
CTR: sod
0.0745
0.0077
9.66
<1e-21
0.0666
CTR: dos
0.0409
0.0045
9.09
<1e-19
0.0240
CTR: dod
0.0015
0.0053
0.29
0.7751
0.0357
size: small
0.0920
0.0173
5.33
<1e-06
0.0312
cardinal: diagonal
0.0204
0.0053
3.88
0.0001
0.0476
CTR: sod & size: small
0.0244
0.0077
3.17
0.0015
CTR: dos & size: small
-0.0054
0.0045
-1.20
0.2317
CTR: dod & size: small
0.0181
0.0053
3.39
0.0007
CTR: sod & cardinal: diagonal
0.0101
0.0028
3.64
0.0003
CTR: dos & cardinal: diagonal
0.0046
0.0037
1.25
0.2125
CTR: dod & cardinal: diagonal
-0.0056
0.0037
-1.51
0.1317
size: small & cardinal: diagonal
0.0042
0.0053
0.79
0.4286
CTR: sod & size: small & cardinal: diagonal
-0.0031
0.0028
-1.12
0.2610
CTR: dos & size: small & cardinal: diagonal
-0.0005
0.0037
-0.12
0.9023
CTR: dod & size: small & cardinal: diagonal
0.0109
0.0037
2.95
0.0032
Residual
0.1904
Model comparison 3
= let = [: m_zcp2, : m_cpx, : m_max]= eval .(nms)= MixedModels.likelihoodratiotest (m_zcp2, m_cpx, m_max)DataFrame (;= nms, = dof .(mods),= round .(deviance .(mods), digits= 0 ),= round .(aic .(mods),digits= 0 ),= round .(aicc .(mods),digits= 0 ),= round .(bic .(mods),digits= 0 ),= vcat (: ., round .(lrt.tests.deviancediff, digits= 0 )),= vcat (: ., round .(lrt.tests.dofdiff, digits= 0 )),= vcat (: ., round .(lrt.tests.pvalues, digits= 3 ))end
1
m_zcp2
23
-24558.0
-24512.0
-24512.0
-24308.0
.
.
.
2
m_cpx
38
-24615.0
-24539.0
-24539.0
-24201.0
57.0
15.0
0.0
3
m_max
153
-24691.0
-24385.0
-24384.0
-23025.0
76.0
115.0
0.998
Other checks
1-element Vector{LinearAlgebra.LowerTriangular{Float64, Matrix{Float64}}}:
[0.6203576608179638 0.0 … 0.0 0.0; 0.22119994699720608 0.26678153140929256 … 0.0 0.0; … ; -0.24965193868875257 0.087476294698932 … 0.4357585158840971 0.0; 0.011471533099315563 -0.0255708216618361 … 0.02800749689407972 0.2461637872603555]
Diagnostic plots of LMM residuals
Do model residuals meet LMM assumptions? Classic plots are
Residual over fitted
Quantiles of model residuals over theoretical quantiles of normal distribution
Residual-over-fitted plot
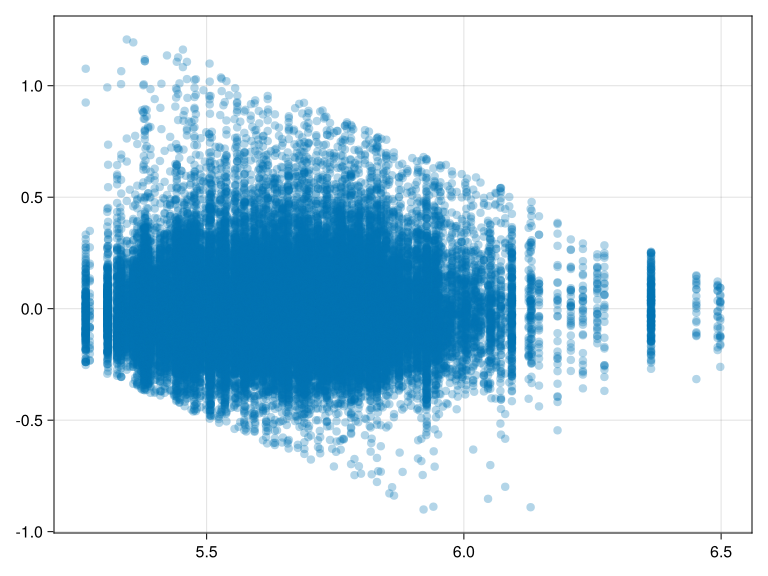
The slant in residuals show a lower and upper boundary of reaction times, that is we have have too few short and too few long residuals. Not ideal, but at least width of the residual band looks similar across the fitted values, that is there is no evidence for heteroskedasticity.
Code
activate! (; type = "png" )scatter (fitted (m_prm1), residuals (m_prm1); alpha= 0.3 )
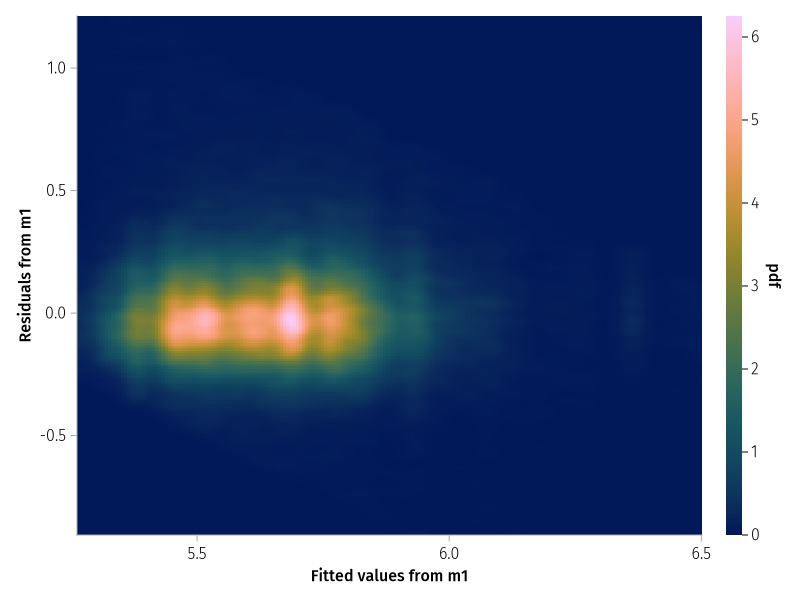
With many observations the scatterplot is not that informative. Contour plots or heatmaps may be an alternative.
Code
set_aog_theme! ()draw (data ((; f= fitted (m_prm1), r= residuals (m_prm1))) * mapping (: f => "Fitted values from m1" , : r => "Residuals from m1" * density ();
Q-Q plot
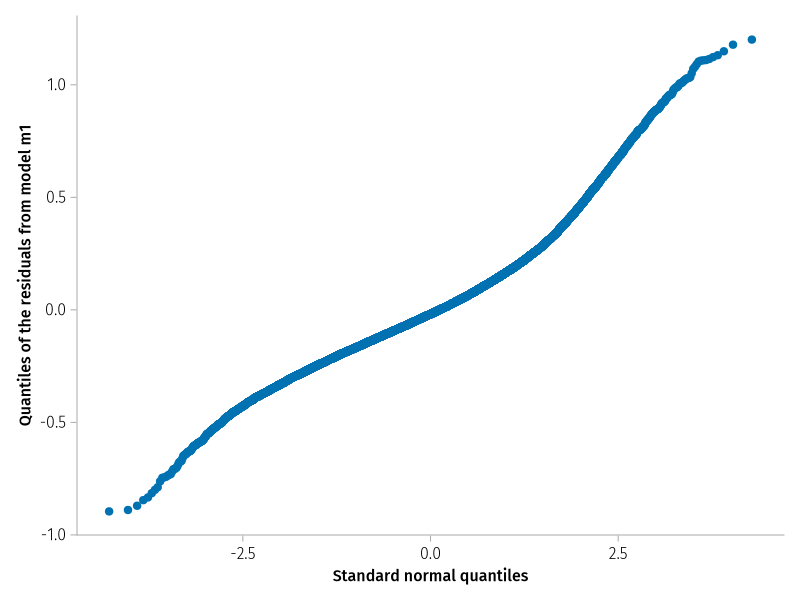
The plot of quantiles of model residuals over corresponding quantiles of the normal distribution should yield a straight line along the main diagonal.
Code
activate! (; type = "png" )qqnorm (residuals (m_prm1);=: none,= (;= "Standard normal quantiles" ,= "Quantiles of the residuals from model m1" ,
Observed and theoretical normal distribution
******We****** can see this in this plot. Overall, it does not look too bad.
Code
activate! (; type = "svg" )let = nrow (dat)= (;= vcat (residuals (m_prm1) ./ std (residuals (m_prm1)), randn (n)),= repeat (["residual" , "normal" ]; inner= n),draw (data (dat_rz) * mapping (: value; color=: curve) * density (; bandwidth= 0.1 );end
Conditional modes
Caterpillar plot
Code
= only (ranefinfo (m_prm1))caterpillar! (Figure (; resolution= (800 , 1200 )), cm1; orderby= 2 )
Parametric bootstrap
Here we
generate a bootstrap sample
compute shortest covergage intervals for the LMM parameters
plot densities of bootstrapped parameter estimates for residual, fixed effects, variance components, and correlation parameters
Generate a bootstrap sample
We generate 2500 samples for the 15 model parameters (4 fixed effect, 7 VCs, 15 CPs, and 1 residual).
= parametricbootstrap (MersenneTwister (1234321 ), 2500 , m_prm1;= (; ftol_rel= 1e-8 ));
Table with 60 columns and 2500 rows:
obj β01 β02 β03 β04 β05 ⋯
┌────────────────────────────────────────────────────────────────────
1 │ -25123.8 5.68038 0.068683 0.039261 -0.00165125 0.0793335 ⋯
2 │ -24465.9 5.66021 0.0667158 0.0418969 -0.00305185 0.100958 ⋯
3 │ -24446.4 5.6924 0.0778707 0.0369443 0.00797653 0.100114 ⋯
4 │ -24617.7 5.69562 0.0766171 0.0463011 -0.00297233 0.0639383 ⋯
5 │ -25167.8 5.68415 0.0684032 0.0385817 0.00248626 0.118136 ⋯
6 │ -24727.8 5.69527 0.0843717 0.0442493 0.00193816 0.0936532 ⋯
7 │ -24512.0 5.67742 0.0707211 0.0418414 -0.00641572 0.076351 ⋯
8 │ -24798.9 5.69362 0.0765456 0.0458011 0.00328823 0.084062 ⋯
9 │ -24392.5 5.68477 0.0818642 0.0496969 -0.000832281 0.0977178 ⋯
10 │ -24585.7 5.68801 0.0796325 0.0344963 0.00629559 0.081441 ⋯
11 │ -25078.8 5.66697 0.0663796 0.0445354 -0.000736944 0.0943102 ⋯
12 │ -24348.5 5.67785 0.0698993 0.0446523 -0.000859312 0.102561 ⋯
13 │ -24973.2 5.68851 0.0732837 0.0421743 -0.00279674 0.0830709 ⋯
14 │ -24617.9 5.70183 0.07733 0.0433379 -0.00455447 0.0694739 ⋯
15 │ -24775.8 5.68347 0.0825422 0.0371819 0.00388279 0.0889206 ⋯
16 │ -24628.0 5.69649 0.0718965 0.0408693 0.000221452 0.0878074 ⋯
17 │ -24397.7 5.70743 0.0966594 0.0363991 0.00133149 0.087301 ⋯
18 │ -24836.3 5.73132 0.0753929 0.0424585 0.00735246 0.0482915 ⋯
19 │ -24806.7 5.68484 0.0744908 0.0369567 0.0060191 0.0529119 ⋯
20 │ -24234.2 5.6857 0.0691212 0.0481745 -6.76522e-5 0.0718787 ⋯
21 │ -24927.4 5.67965 0.066628 0.0441619 0.000997131 0.0939013 ⋯
22 │ -24044.7 5.68194 0.0675943 0.0364103 0.0108015 0.096463 ⋯
23 │ -24804.0 5.72333 0.0826845 0.0393307 0.00877465 0.0955252 ⋯
⋮ │ ⋮ ⋮ ⋮ ⋮ ⋮ ⋮ ⋱
Shortest coverage interval
DictTable with 2 columns and 38 rows:
par lower upper
────┬────────────────────────
β01 │ 5.65966 5.72791
β02 │ 0.0602683 0.0902347
β03 │ 0.0326438 0.0490006
β04 │ -0.00832038 0.0114628
β05 │ 0.0585181 0.124489
β06 │ 0.00932821 0.030229
β07 │ 0.00994937 0.0391542
β08 │ -0.0138358 0.00317468
β09 │ 0.00770406 0.0274789
β10 │ 0.00485596 0.0157058
β11 │ -0.00249685 0.0117935
β12 │ -0.0124823 0.00202088
β13 │ -0.00582681 0.0137215
β14 │ -0.00828443 0.00227533
β15 │ -0.00842329 0.00599236
β16 │ 0.00369609 0.0179879
ρ01 │ 0.380561 1.0
ρ02 │ -0.342485 0.999999
ρ03 │ -0.685044 0.682981
ρ04 │ 0.466487 0.989877
ρ05 │ 0.330399 0.883748
ρ06 │ -0.273379 0.932581
ρ07 │ -0.729693 -0.0549567
⋮ │ ⋮ ⋮
We can also visualize the shortest coverage intervals for fixed effects with the ridgeplot() command:
Code
ridgeplot (samp; show_intercept= false )
Comparative density plots of bootstrapped parameter estimates
Residual
Code
draw (data (tbl) * mapping (: σ => "Residual" ) * density ();= (; resolution= (800 , 400 )),
Fixed effects and associated variance components (w/o GM)
The shortest coverage interval for the GM ranges from x to x ms and the associate variance component from .x to .x. To keep the plot range small we do not include their densities here.
Code
= renamer (["(Intercept)" => "GM" ,"CTR: sod" => "spatial effect" ,"CTR: dos" => "object effect" ,"CTR: dod" => "attraction effect" ,"(Intercept), CTR: sod" => "GM, spatial" ,"(Intercept), CTR: dos" => "GM, object" ,"CTR: sod, CTR: dos" => "spatial, object" ,"(Intercept), CTR: dod" => "GM, attraction" ,"CTR: sod, CTR: dod" => "spatial, attraction" ,"CTR: dos, CTR: dod" => "object, attraction" ,draw (data (tbl) * mapping (: β02 , : β03 , : β04 ] .=> "Experimental effect size [ms]" ;= dims (1 ) => renamer (["spatial effect" , "object effect" , "attraction effect" ]) => "Experimental effects" ,* density ();= (; resolution= (800 , 350 )),
The densitiies correspond nicely with the shortest coverage intervals.
Code
draw (data (tbl) * mapping (: σ2 , : σ3 , : σ4 ] .=> "Standard deviations [ms]" ;= dims (1 ) => renamer (["spatial effect" , "object effect" , "attraction effect" ]) => "Variance components" ,* density ();= (; resolution= (800 , 350 )),
The VC are all very nicely defined.
Correlation parameters (CPs)
Code
draw (data (tbl) * mapping (: ρ01 , : ρ02 , : ρ03 , : ρ04 , : ρ05 , : ρ06 ] .=> "Correlation" ;= dims (1 ) => renamer (["GM, spatial" , "GM, object" , "spatial, object" ,"GM, attraction" , "spatial, attraction" , "object, attraction" ]) => "Correlation parameters" ,* density ();= (; resolution= (800 , 350 )),
Three CPs stand out positively, the correlation between GM and the spatial effect, GM and attraction effect, and the correlation between spatial and attraction effects. The second CP was positive, but not significant in the first study. The third CP replicates a CP that was judged questionable in script kwdyz11.jl.
The three remaining CPs are not well defined for log-transformed reaction times; they only fit noise and should be removed. It is also possible that fitting the complex experimental design (including target size and rectangle orientation) will lead to more acceptable estimates. The corresponding plot based on LMM m1_rt for raw reaction times still shows them with very wide distributions, but acceptable.
References
Baayen, H., Vasishth, S., Kliegl, R., & Bates, D. (2017). The cave of shadows: Addressing the human factor with generalized additive mixed models.
Journal of Memory and Language ,
94 , 206–234.
https://doi.org/10.1016/j.jml.2016.11.006
Bates, D., Kliegl, R., Vasishth, S., & Baayen, H. (2015).
Parsimonious mixed models . arXiv.
https://doi.org/10.48550/ARXIV.1506.04967
Kliegl, R., Kushela, J., & Laubrock, J. (2015). Object orientation and target size modulate the speed of visual attention . Department of Psychology, University of Potsdam.
Kliegl, R., Wei, P., Dambacher, M., Yan, M., & Zhou, X. (2011). Experimental effects and individual differences in linear mixed models: Estimating the relationship between spatial, object, and attraction effects in visual attention.
Frontiers in Psychology .
https://doi.org/10.3389/fpsyg.2010.00238